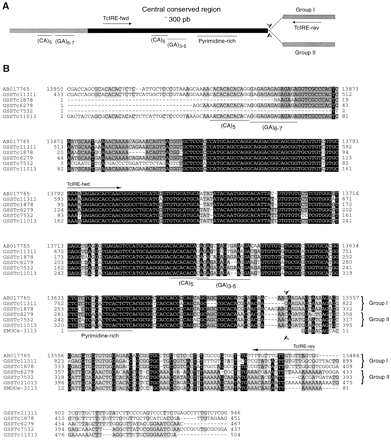
Structure of TcIRE. (A) General scheme of the structure of TcIRE. (B) Five GSS containing a copy of TcIRE were aligned using CLUSTALW with the corresponding region of the 25-Kb cosmid sequenced by Gao et al. 1999 (GenBank accession no. AB017765) and the last portion of the 3′ UTR from the Emuce-31l3 mucin gene. The rest of the mucin gene does not show any homology with the sequences aligned and was cut off for the sake of clarity. Coloring is based on BLOSUM 62 scores: 3.0, black; 1.5, gray; 0.5, light gray. Similar residues are colored as the most conserved one. Arrows indicate the two oligonucleotides used to generate the probe for the Southern blot analysis. Arrowheads indicate the site of divergence between the two groups of TcIRE sequences. The top three sequences, includingAB017765, are representative of one group of sequences, denoted Group I, whereas the other sequences, including the last portion of the 3′-UTR region from the Emuce-31l3 mucin gene are representative of another group, denoted Group II.











