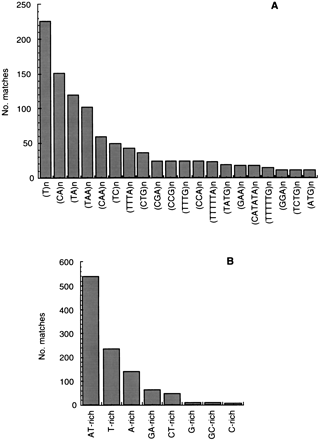
Microsatellite repeats and low-complexity regions in theTrypanosoma cruzi genome. Simple repeats and low-complexity regions were searched for in the T. cruzi GSS database usingREPEATMASKER as described in the text. (A) The 20 most abundant microsatellite repeats in the survey are shown. The minimum value of n is the one that gives a Smith-Waterman (SW) score ⩾180, which is the cutoff to consider a match as positive. This value varied from 18 for a single nucleotide repeat to 3 for a hexanucleotide repeat. Each named microsatellite in the graph includes all combinations thereof; so (A)n also includes its complement (T)n, and (ATG)n also includes (CAT)n, (ATC)n, (TCA)n, (TGA)n and (GAT)n. (B) Low-complexity regions were searched for as described in the text. The length of the regions detected varied from 16 bp to 308 bp.











