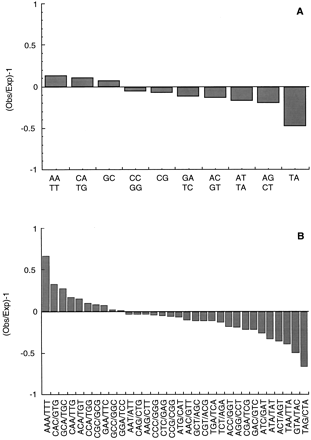
Frequency of di- and tri-nucleotide repeats in the Trypanosoma cruzi genome. The total of 11,459 sequences were used to search for the occurrence of all possible words of length 2 (A) and 3 (B) on both strands of the sequences usingCOMPSEQ. The expected frequency of each word is based on the assumption that all words have the same probability of occurrence. Di- and tri-nucleotide frequencies are expressed as Observed (Obs)/Expected (Exp) − 1, so that negative values correspond to suppressed di- and tri-nucleotides and positive values correspond to di- and tri-nucleotides with frequencies over that expected. Because the search is done on both strands, only one reverse complementary di- or tri-nucleotide of a pair is shown.











