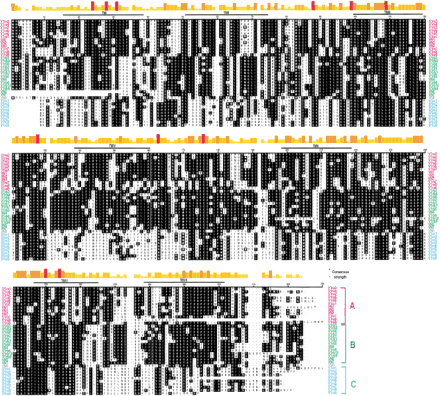
Alignment of predicted amino acid sequences of mouse and rat V1r genes. Predicted amino acid sequences from V1r sequences were aligned with the other mouse (V1ra2, V1rb5, and V1rb6) and rat (VN1, VN2, VN3, VN4, VN5, VN6, and VN7) V1r sequences available in public databases. The alignment was performed using the Clustal V method of the MegAlign program. The V1r sequences were aligned by the first methionine common to all open reading frames. Predicted consensus positions of TM domains are indicated as TM I-VII. Consensus residues are shown as white lettering on black background. Consensus strength is shown ontop. The shortest, yellow bars represents the least conserved amino acids; the tallest, red bars represent amino acids that are common to all sequences. V1r genes from groups a, b, and c are indicated in pink, green, and blue, respectively. VN6 from rat does not belong to any of the three groups and is indicated in gray. The publicly available sequences of VN5 and VN7 from rat seem to be incomplete, precluding full-length alignment.











