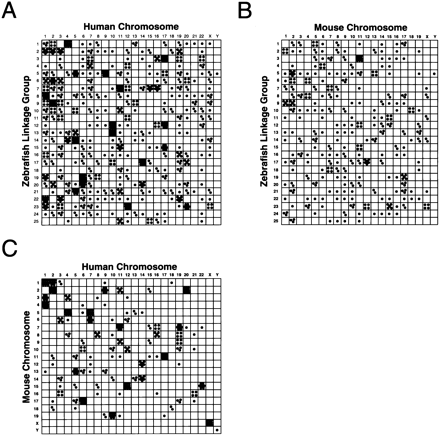
Oxford grids showing conservation of synteny among zebrafish, human, and mouse. Each dot represents an orthologous gene pair plotted by position in the two species compared. Boxes that contain more than one dot represent conserved syntenies. (A) Zebrafish–human comparison; (B) Zebrafish–mouse comparison; (C) Human–mouse comparison. Human and mouse orthologs of zebrafish genes mapped in the heat shock panel are listed in Table 1. Orthologs of genes mapped in previous work (Postlethwait et al. 1998;Gates et al. 1999) are also included in these grids. The genes in the zebrafish–human grid are summarized in Table 2. All genes in aHox complex are represented by a single dot. All three grids clearly show a nonrandom distribution; χ2 values, calculated according to Gates et al. (1999) are: χ2 = 158, zebrafish–human; χ2 = 26.0, zebrafish–mouse; and χ2 = 220, human–mouse. For 1 degree of freedom, a χ2 >10.83 indicates a significant difference between a random distribution and the observed distribution at a significance level P <0.001. The map positions of zebrafish genes and expressed sequence tags summarized in the graphs were derived from this study and from previous work (Amores et al. 1998; Postlethwait et al. 1998; Gates et al. 1999; Kelly et al. 2000).











