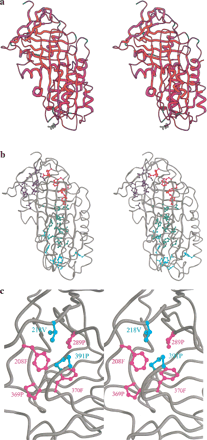
Amino acid conservation in the serpin superfamily. (A) Kabat variability in residues appearing at each site, mapped onto the structure of cleaved α1-antitrypsin. The color scheme ranges from red (low variability) to blue (high variability). Residues corresponding to positions in which >20% of sequences contain gaps are shown in green. The figure was produced using MOLSCRIPT (Kraulis 1991). (B) Cleaved α1-antitrypsin indicating residues conserved in >70% of sequences in ball and stick representation. Residues are colored according to the functional region of the serpin in which they are found: (blue) gate; (red) breach; (green) shutter. Residues outside these regions are in cyan. (C) Packing of conserved residues within the gate region. Phe208, Pro289, Pro369, and Phe370 are almost invariant (conserved in >95% of sequences) and are colored magenta. Two other highly conserved residues—Val218 and Pro391—are colored cyan.











