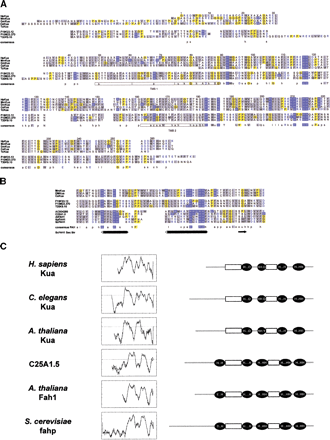
Kua, a new family of conserved proteins. (A) Alignment of Kua proteins predicted from PCR-generated cDNAs (Homo sapiens andDrosophila melanogaster), ESTs (Mus musculus andT. cruzi), or genomic sequences (Caenorhabditis elegans and Arabidopsis thaliana, F19K23.12, F10M23.370, and T20K9.10). The prediction for the A. thaliana F10M23.370 genomic sequence is supported by EST AA712589. Color codes: blue background, His; yellow background, Gli and Pro; grey background, hydrophobic residues; blue, charged residues. Key for consensus sequences: a, aromatic (F, H, W, Y); c, charged (D, E, H, K, R); h, hydrophobic (F, L, M, V, W, Y, I); l, aliphatic (I, L, V); o, alcohol (S, T); p, polar (C, D, E, H, K, N, Q, R, S, T); s, small (A, C, D, G, N, P, S, T); u, tiny (A, G, S); x, helix breaking (G, P);−, negative (D, E); +, positive (H, K, R). Potential transmembrane domains are boxed in the consensus sequence. (B) Alignment of the segments of proteins containing the two histidine-rich motifs detected by PHI–BLAST search of GenBank nr database with the profile H-x-(YWF)-x-H-x(8,25)-(RK)-x(2)-H-x(2)-H-H, generated after the consensus sequence for Kua proteins in this region. Shown are fatty acid hydroxylases from D. melanogaster (AC004280) C. elegans (C25A1.5), A. thaliana (AtFAH1),Schizosaccharomyces pombe (SpFAH1) and Saccharomyces cerevisiae (ScFAH1). (Bottom) Secondary structure predictions for yeast FAH (cylinders, alpha helix; arrow, beta sheet). (C) Transmembrane domain predictions and compared topological models for Kua proteins and fatty acid hydroxylases. Leftpanel, predictions of transmembrane domains for human (AF155120), worm (Y53C10A.5), and plant (F19K23.12), Kua and worm (C25A1.5), and plant (At Fah1) fatty acid hydroxylases. Right panel, diagrams representing the positions of the histidine-rich motifs in the same proteins, relative to the predicted transmembrane domains. Diagrams are not drawn to scale.











