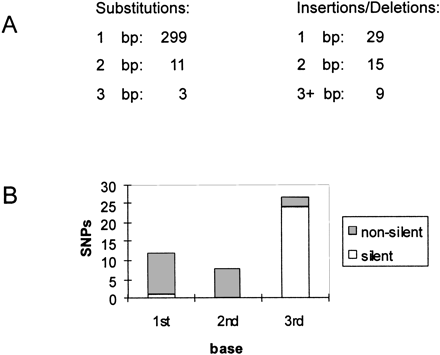
Nature of the SNPs. (A) The number of base pairs involved in substitutions, insertions, and deletions is indicated. Note that we find on average one SNP per 2000 bp. After checking the Bristol N2 sequence for 63 potential SNPs, we found 3 to be a sequence error in N2. Thus, we estimate the error rate of the C. elegansgenome sequence to be ∼1 in 42,000. (B) The number of SNPs involved in the first, second, or third base of a codon in coding sequences is plotted. It also shows whether the SNP alters the amino acid. Coding sequences were predicted by Genefinder.











