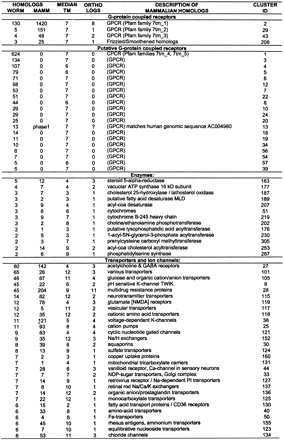
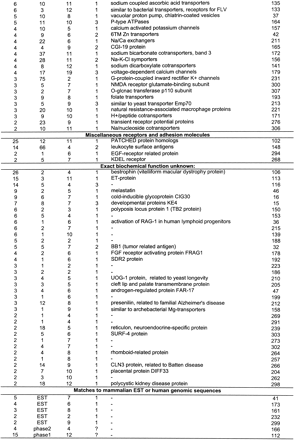
Description of Clustered Worm Protein Families
|
-
The columns WORM and MAMMALIAN show the number ofCaenorhabditis elegans and mammalian homologs found in used protein databases with each HMM-model (with HMMPFAM cutoff level E < 1e-2). The column MEDIAN TM shows median number of predicted transmembrane domains in all matching worm proteins. The number of mammalian–worm ortholog assignments within each cluster is shown in column ORTHOLOGS. The description line shows description of mammalian homologs. Each orthology assignment is described in detail in Table 2, available as supplementary information at http://www.genome.org. Families are organized into groups by their general biochemical function. Fifty-six families for which mammalian homologs were not found are not shown here. Mammalian homologs found in unfinished genomic DNA are marked “phase1” or “phase2”. This table can also be downloaded from ftp.cgr.ki.se/pub/data/worm.











