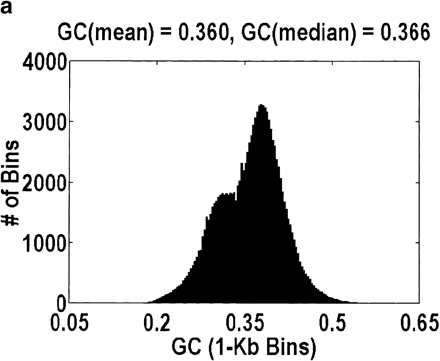
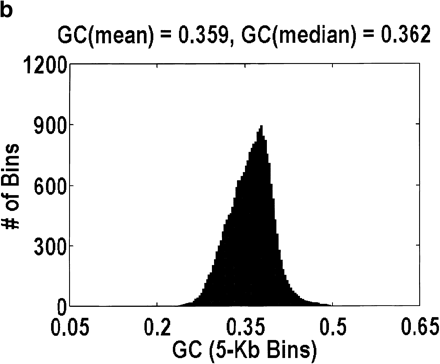
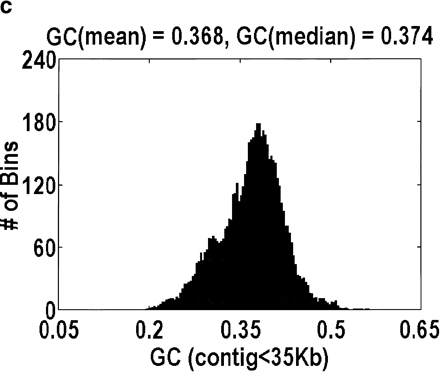
Distribution of GC content for anonymous genomic sequence inArabidopsis thaliana. The idea that a significant fraction of the genome is intergenic, coupled with the fact that intergenic DNA has a lower GC content than intragenic DNA, suggests that this distribution will be bimodal. However, the bimodality is easily obscured by how the data are plotted. a and b differ in the size of the bins over which the GC content is computed, 1 kb and 5 kb, respectively. Bin sizes larger than the average gene size of 2.6 kb obscure the effect because every bin is likely to contain a mixture of intragenic and intergenic DNA. a and c differ in the genomic contigs that are plotted (every contig or only contigs <35 kb, respectively). By removing the large-insert clones favored by the genome centers, what is left behind are those sequences that were analyzed only because they contain a likely gene. Hence, the bimodality disappears.











