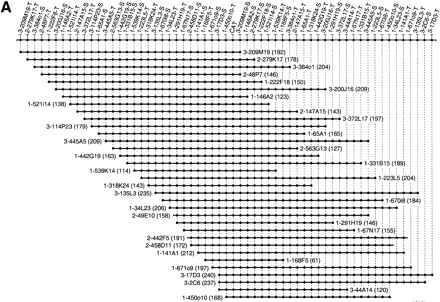
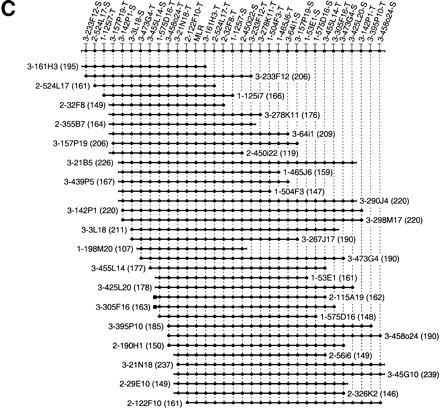
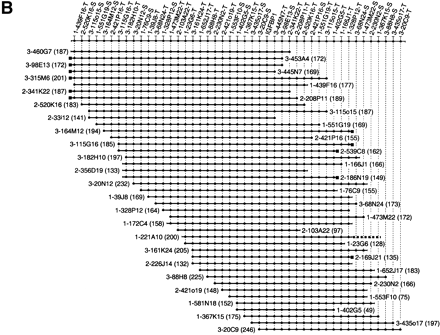
Three 400-kb high-resolution end probe-based BAC–PAC contigs in murine CAT (A), IGFBP1 (B), and MLR (C) gene regions. The contigs have been assembled according to the hybridization results using SEGMAP contig assembly software. The deduced markers are depicted along the top and each short horizontal line with black circles represents PAC or BAC clones. The clone names are represented by the library name, plate number, and well position, e.g., 3-209M19 stands for RPCI-23 library, plate-209, M19 well. RPCI-21, RPCI-22, and RPCI-23 are shortened to 1, 2, and 3. The T7 and SP6 end markers are condensed as -T and -S after the clone name. The size of each clone is indicated in kb in parenthesis. Because the markers are spaced evenly in the contigs, the length of the horizontal line does not represent the clone size accurately. The dotted line in the IGFBP1 contig (B) indicates the partly chimeric clone 1-221A10. The black squares in B and C represent an identical clone end among neighboring clones. A unique marker was not designed from these ends because of the presence of repetitive sequence. However, these end sequences were used to ascertain additional 10 nonchimeric clones.











